DNA Fingerprinting | Biology Science Fair Project
DNA Fingerprinting | Biology Science Fair Project
DNA fingerprinting is a powerful new forensic technology, that many argue is the greatest tool in the history of forensic science. But as is often the case for new technologies, its acceptance by society was not straightforward.
This project investigates this technology describing how it is done, its uses, and its indirect path of acceptance in the courtoom.
DNA FINGERPRINTING TYPES
DNA fingerprinting is one of the greatest identification systems we have to recognize an individual or living organism. Every living creature is genetically different in its own way, except for identical twins, triplets etc. DNA is comparable to a serial number for living things. Each individual contains a unique sequence that is specific to that one organism. Unlike traditional fingerprints which can be surgically altered or self mutilated, the DNA sequence can not easily be changed once the material is left at a crime scene, thus increasing its effective use in forensics, and the probability of finding an exact match. This method of identification is useful in many applications such as forensics, paternity testing, and molecular archeology, which we will discuss later on in this chapter. To further understand DNA fingerprinting we must first discuss the basics of DNA.
Introduction to DNA Basics
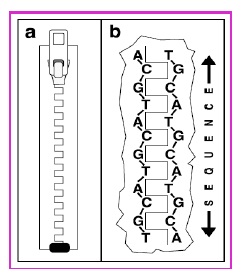
DNA, also known as deoxyribonucleic acid, contains a specific sequence of bases called nucleotides which contain the information of all the characteristics of living organisms. This information was inherited through the DNA of their parents. DNA is found in almost every cell of every living organism. The DNA represents the “instruction book” for making living organisms. The four nucleotides that constitute the sequences of DNA are adenine (A) which bonds exclusively with thymine (T), and guanine (G) which bonds exclusively with cytosine (C). The molecular structure of DNA can be imagined as a zipper (Figure-1) with each tooth representing one of the four letters (A, C, G, or T) and with opposite teeth forming either of the two pairs, AT or GC.
RFLPs and VNTRs
A chromosome is the visible state of genetic material during the division phase of a cell. Humans have 23 pairs of chromosomes, which makes 46 individual chromosomes. Half of the chromosomes of an individual come from the mother and the other half from the father. Chromosomes are found in the nucleus, and contain a linear strand of DNA. The DNA molecule is twisted onto itself and the super-coiled molecule is enclosed in proteins which help maintain its shape. The chromosomes carry the genes that make each individual.
Now that we have a better understanding of what DNA actually is, lets move on to the basics of making a DNA fingerprint. There are three types of DNA fingerprints: RFLPs, VNTRs, and STRs. Restriction fragment length polymorphisms, or RFLPs as they are commonly known, were the first type of DNA fingerprinting which came onto the scene in the mid- 1980’s. RFLP’s focus on the size differences of certain genetic locations.
The first step in creating an RFLP fingerprint is obtaining and isolating the DNA. DNA can be obtained from almost any of the cells or tissues in the human body. You do not need a large amount of tissue or blood to provide enough DNA for analysis. The DNA is then extracted from the blood or tissue sample, and from here we carry out our second step in the process which is the cutting, sizing, and sorting of the DNA sample. DNA is cut using restriction enzymes, which cut the DNA stand at specific places. Restriction enzymes are usually isolated from bacteria that use them to degrade foreign DNA like viral DNA. Each type of restriction enzyme recognizes and cuts a particular DNA sequence.
The DNA at this point is cut into a various array of pieces which are sorted according by size through a process called electrophoresis. In this process the DNA particles are mixed into a buffer solution and applied to a gel made from seaweed agarose. Each side of the gel is connected to an electrical current. The DNA is negatively charged due to its phosphate groups, so it migrates towards the positive electrode or anode. The smaller pieces of DNA move faster (sieve) through the gel than the larger ones, so this provides the basis of the fragment separation. “This technique is the DNA equivalent of screening sand through a progressively finer mesh screens to determine particle sizes” (Betsch, 2005).
The band pattern that the DNA creates in the agarose gel is then transferred to a nylon sheet. To complete this transfer a nylon sheet is placed on the gel and left to soak overnight in a high salt solution. After the soaking procedure is completed, the nylon membrane contains the same pattern of DNA as occurred in the original gel. The membrane is now prepared to undergo its probing phase. Radioactive or fluorescently labeled probes are hybridized onto the nylon membrane, which bind to specific DNA sequences present in the pattern to produce a pattern of bands which create the DNA fingerprint. This process can be performed with several different probes simultaneously to make the final product which looks very similar to the bar codes you see in retail stores. Figure 2 shows an actual RFLP-type DNA fingerprint.
Variable number tandem repeats, or VNTRs represent specific locations on a chromosome in which tandem repeats of 9-80 or more bases repeat a different number of times between individuals. These regions of DNA are readily analyzed using the RFLP approach and a probe specific to a VNTR locus. The fragments are a little shorter than RFLPs (about 1-2 kilo base pairs), but are created through the exact same process. Figure 3 shows an example of a VNTR fingerprint
Since RFLPs and VNTRs are created in the same fashion, they exhibit the same overall advantages and disadvantages. Some of the advantages of these types of DNA fingerprints are that they are the most stable and reproducible, which is a valuable trait to have when you are trying to determine an exact match of a person’s DNA, which must exclude billions of other people’s DNA with a certain degree of confidence. They are also easier to prevent contamination since the DNA sample is larger than with other types of DNA fingerprints, and small amounts of DNA contamination does not alter the analysis. Some of the disadvantages of RFLPs and VNTRs include they are very time consuming (especially the probe hybridization step), relatively large amounts of DNA must be used to obtain an adequate sample, too many polymorphisms may be present for a short probe, and the cost is very high due to labor and time requirements
STRs and PCR
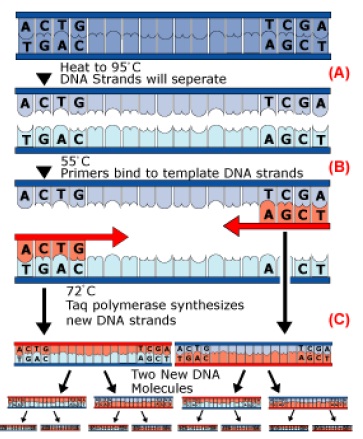
Currently, the most popular method of DNA fingerprinting are short tandem repeats, or STRs for short. Unlike VNTRs which analyze minisatellites that have repeat sequences of 9-80 base pairs, STRs use microsatellites which have repeat sequences of only 2-5 base pairs, introducing the “less is more” philosophy to the world of DNA fingerprinting. This was a big step forward in forensic science since the length of DNA fragment being analyzed is short enough to be amplified by polymerase chain reaction (PCR), so now we are able to analyze a very small sample of DNA that is quicker and easier than any previously known method and match it to a person’s identity. PCR was developed in the mid 1980’s and used the same principles that cells use to replicate DNA to amplify the specified region, which is usually between 150-3,000 base pairs in length. In order to amplify the DNA sequence, a pair of short priming sequences (which are complimentary to the ends of the targeted sequence), a special heat-resistant DNA polymerase called Taq polymerase, and a solution of the four DNA bases are all mixed together in a test tube which contains a few copies of the targeted DNA sequence (Genetic Analysis, 2004).
The DNA is then amplified (or replicated) by the repetition of a cycle which contains three vital steps:
• The solution is heated to 95°C to unzip the double helix DNA structure (Fig. 4A).
• The solution is cooled to 55°C to allow the primers to bind to the ends of the DNA (Fig-4B).
• The solution is then reheated to 75°C which is the optimal temperature for the Taq polymerase to create new copies of each DNA strand
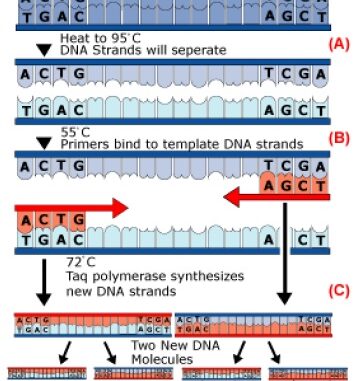
One PCR cycle takes approximately 2 minutes to complete. Each cycle doubles the amount of the previous amount of targeted sequences in the test tube, so it only takes about 50 cycles to produce hundreds of thousands of DNA copies So long as primers are chosen to flank an STR site, the band amplified will represent the STR locus, and a simple gel or column will determine the band length.
Thus this procedure avoids the lengthy probe hybridization step to membrane of the RFLP/VNTR approaches. STRs are currently the most popular type of DNA fingerprint, since the whole PCR process takes only a few hours, compared to RFLP/VNTR probe hybridization and film exposure which can take several days. STRs can use much smaller samples of DNA than RFLPs/VNTRs, and can even use partially degraded DNA to create a fingerprint. Thus, the integrity and quality of the DNA sample is not as great a factor with STRs than with the traditional methods of DNA fingerprinting (Introduction to STRs, 2005). The current standard forensic protocol analyses 13 core STR loci which have been carefully chosen for their uniqueness. The only disadvantage of the STR approach is it is sensitive to contaminating DNA, so usually the STR approach is used first, followed by a VNTR analysis if contamination is suspected, and enough DNA is available.
Applications of DNA Fingerprinting
DNA fingerprinting is used in a variety of applications all over the world. They can be used to solve criminal cases such as rape, used to conduct a paternity test, or even used to determine the authenticity of rare sports memorabilia. Whatever the case, it is evident that DNA fingerprinting has revolutionized the way the world identifies biological matches. We will discuss a few examples of these applications and their importance below
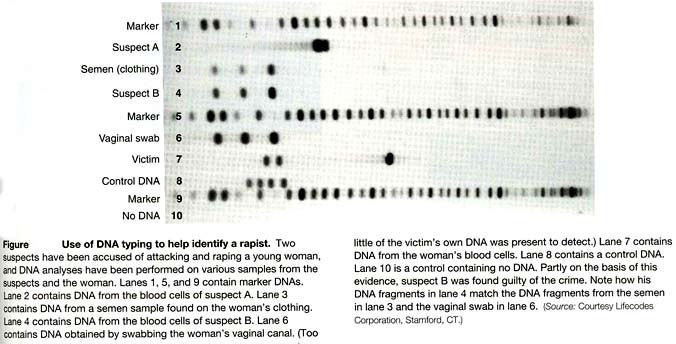
One of the first accepted uses of DNA fingerprinting was in the investigation of sexual assault and rape cases. Detectives only had to match the DNA of the semen found at the scene of the crime with the DNA of any potential suspect to determine who was guilty of committed the crime. A DNA sample from the rapist could be obtained from a simple vaginal swab from the victim or any other semen that was released in the area during the assault. The figure-6 below shows how a DNA fingerprint can help determine who is guilty of a sexual assault.

As seen from figure-5, suspect B (lane 4) is guilty of rape because his DNA fragments match that of the semen found on the victim’s clothes (lane 3) and also in the vagina (lane 6). Suspect A (lane 2) is clearly not the rapist because his DNA fragments do not match the semen found on the victim’s clothes or the semen from the vaginal swab. DNA fingerprinting is very useful in such an application because it provides the police with an exact match of who left evidence at the crimescene.
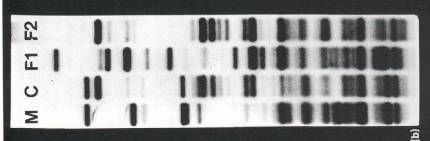
Paternity tests are another application of DNA fingerprinting that has been incorporated around the world. In paternity tests potential fathers of the child have their DNA analyzed with the child and mother’s DNA in order to see which of the potential fathers has the most DNA in common with the child in question. Figure 6 shows an example of a RFLP used to determine which potential father (F1 and F2) is the real father of the child (C). As you can see in the figure below, the second father tested (F2) seems to have more DNA in common with the child than that of the first father tested (F1).

Another application of DNA fingerprinting is a more recent method in molecular archeology. This method of archeology uses DNA to determine a species of an archeological discovery or to trace blood lines of animal or human remains. DNA may be extracted from biological remains, hair, teeth, body tissues, or even fossils. The best climates to preserve DNA are very cold temperatures and arid climates. Some examples of specimens from these types of climates are the “Tyrolean Ice-Man”, who was found in the Alps, and the mummies of Egypt found in the dry desert. The ice man was found to be around 5300 years old, and DNA was extracted from the remains of his gut which found small traces of food that he ate (Ice Man, 2005). This was one of the most historic archeological discoveries in the last century. DNA fingerprinting is an important tool for archeologists to piece together information that links the past to us today. Figure 7 shows a picture of the “Tyrolean Ice-Man”.
DNA fingerprinting is even used in the world of sports collectibles. With sports collectors spending gigantic amounts of money to own a piece of sports history, there needed to be a way to validate the authenticity of the rare memorabilia. The memorabilia can be treated with a synthetic DNA smear, in which the item is coated with a secret DNA sequence where the original batch of DNA is then destroyed. The collectible can then be auctioned off giving the buyers assurance that the product is indeed authentic. This is just another instance of how DNA fingerprinting can be used in today’s world.
DNA FORENSICS
Forensic science is the art of piecing together a crime scene in order to determine how the crime was committed and who was responsible. DNA evidence is one of the most prominent pieces of evidence that is used in the United States judicial system today. Just because techniques exist that allow DNA to be analyzed at a crime scene does not necessarily mean that evidence was collected correctly to avoid contamination, or was stored correctly to prevent DNA degradation. As we will learn in Chapter-3 when we discuss landmark DNA court cases, many times DNA evidence has been prevented from use in a particular trial due to improper handling. The purpose of this chapter is to discuss some of the current knowledge about proper DNA handling.
DNA evidence can be collected by various means from almost any biological sample that was left at the scene of the crime. In the past when someone committed a crime such as a sexual assault, unless there were witnesses there was no real way of proving that a specific person was guilty. Normal blood types are not that exclusive. Now with DNA forensics, a level of certainty can be established that is recognized as valid evidence in a criminal case, either for the prosecution or the defense. There have been numerous instances where men were charged with rape in the past and had DNA analyzed from the crime scene only to find out that they were innocent all along. Figure 1 shows an example of how DNA analysis can help determine who is guilty of the crime in question. Note how the crime scene sample matches suspect 3. We will now discuss the proper techniques to conduct a forensic investigation.